Methylation using Sequencing
Among the different existing methods to study methylation profiles, DNA sequencing is probably the most efficient to date. IntegraGen presents an overview of the most innovative and cost-effective sequencing techniques to guide you in the choice of the best method taking into consideration the type of project, the region of DNA to be explored, the type of samples (tissue or liquid biopsy), the DNA quantity and quality, as well as your budget constraints.
HOW TO CHOOSE THE RIGHT METHYLATION TECHNIQUE?
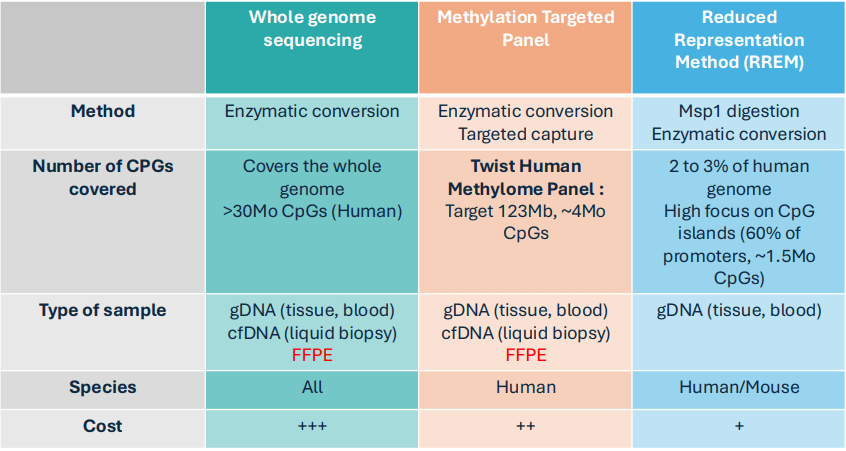
While whole genome sequencing still faces cost constraints, reduced representation sequencing methods cover mostly CpG islands. In addition, by offering DNA capture method, IntegraGen offers the newest and best sequencing technique to study methylation profiles in all the DNA methylated regions that matter (not only CpG islands, but also, shelves, shores, and open sea regions).
OUR EXPERIENCE
- Develop large scale methylation analysis using high throughput sequencing since 2011, and using methylation arrays since 2006.
WHAT MAKES INTEGRAGEN YOUR BEST PARTNER?
- Expertise in project design and analysis plan implementation
- Protocol available for circulating free DNA
- Protocol available for free circulating DNA
- Extraction available from tissues or liquid biopsies
- Bioinformatic analysis available
OUR GUARANTEES
- High-quality data, read number guaranteed
- Conversion efficiency > 99.5%
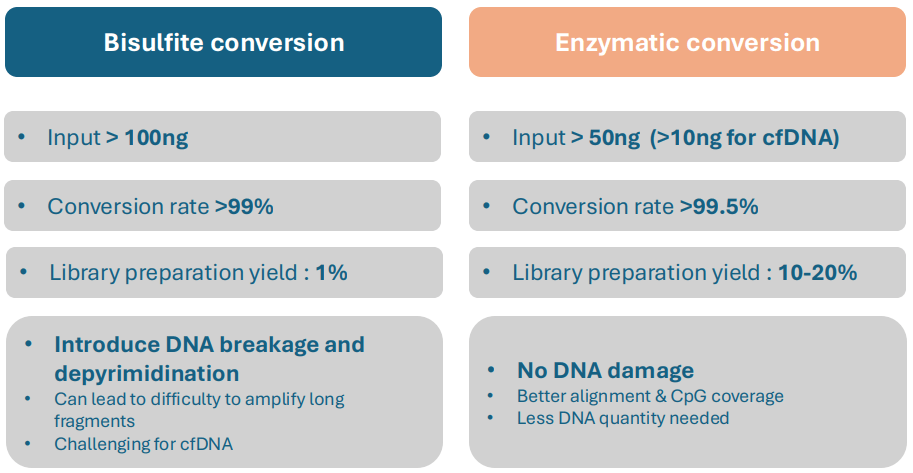
WHY IS ENZYMATIC CONVERSION PREFERED OVER BISULFITE CONVERSION?
Although the sodium bisulfite conversion of DNA is considered as the gold standard for DNA methylation, this method has some biases compared to enzymatic conversion. Indeed, the chemical reaction with sodium bisulfite damages and degrades the DNA, leading to its fragmentation and loss. The resulting reduced DNA quantity will further bias sequencing data by affecting the DNA quality and CpG coverage. The enzymatic conversion method chosen by IntegraGen leads to less DNA damage, ensuring higher DNA quality and yield, as well as a greater conversion efficiency (higher CpG coverage and alignment quality). This technique is entirely appropriate for the methylome analysis of samples containing a low amount of starting DNA (e.g. cfDNA), amplification of long DNA fragments, and is applicable to DNA capture method.